-
Recursion Announces the Release of OpenPhenom-S/16 in Google Cloud’s Model Garden
Source: Nasdaq GlobeNewswire / 12 Nov 2024 16:54:03 America/New_York
SALT LAKE CITY, Nov. 12, 2024 (GLOBE NEWSWIRE) -- Recursion (NASDAQ: RXRX), a leading clinical stage TechBio company decoding biology to industrialize drug discovery, announced today the release of OpenPhenom-S/16, a publicly available foundation model in Google Cloud’s Vertex AI Model Garden. This will enable Life Science companies to use OpenPhenom-S/16 on Google Cloud when building apps that require microscopy data to assess the effectiveness of drug treatments, analyze cultures, study tissues and ultimately accelerate drug discovery and for other uses.
“We are thrilled to announce that Recursion is setting a new gold standard for the industry with the release of OpenPhenom-S/16, a foundation model for microscopy data trained using masked-autoencoder-based self-supervised learning on more than three million microscopy images from publicly accessible datasets (RxRx3 and JUMP-CP),” said Ben Mabey, chief technology officer at Recursion.
Large-scale cell microscopy assays have become instrumental in unveiling novel biological processes, potential drug candidates, and targets. Traditionally, laboratories have relied on legacy image segmentation and feature extraction software packages that need to be tuned for each new assay and are often brittle to minor changes in assay conditions. With the increasing scale and complexity of private and public datasets, such as RxRx3 and JUMP-CP, these existing software packages and associated downstream analysis methods struggle to handle the sheer volume of data effectively.
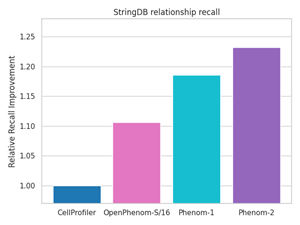
OpenPhenom-S/16 will allow non-commercial researchers to potentially replace their existing workflows with this off-the-shelf model that outperforms traditional microscopy analysis pipelines without requiring any additional tuning or training. For example, with OpenPhenom-S/16, Recursion presents an unprecedented improvement in recalling known biological relationships from the StringDB database in the public JUMP-CP cpg0016 dataset.
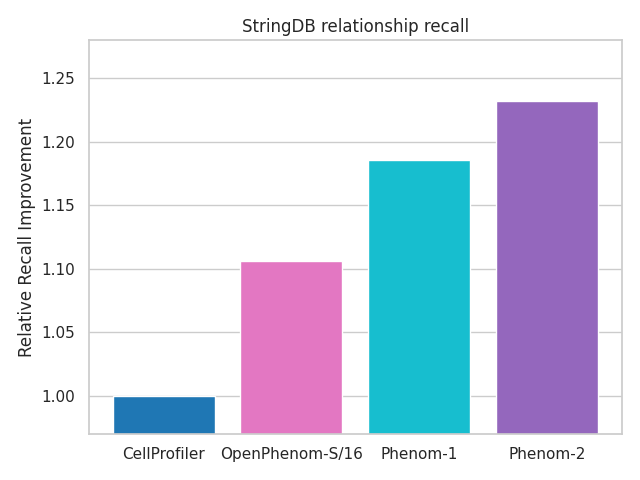
Figure 1 demonstrates the improvement in recalling known biological relationships (PLoS Comp Bio 2024) from the StringDB database in the public JUMP-CP dataset when using OpenPhenom-S/16 embeddings compared to the gold standard approach with CellProfiler.
"We’re pleased Recursion is bringing OpenPhenom-S/16 to the life science community through Google Cloud's Vertex AI Model Garden,” said Shweta Maniar, global director of Life Science Strategy & Solutions, Google Cloud. “This powerful foundation model, combined with the scalability and accessibility of our Vertex AI platform, has the potential to accelerate microscopy analysis by automating classification of cellular structures, detecting patterns and anomalies, and extracting valuable insights from complex microscopy data. By democratizing access to this cutting-edge technology, Recursion will empower researchers around the globe to make new discoveries and advance the field of biomedicine."
As a leader in this space, Recursion also has models for commercial use for its own pipeline and partnership programs. Recursion has featured the performance of two generations of proprietary models trained on Recursion’s internal microscopy, Phenom-1 (CVPR 2024) and Phenom-2 (NeurIPS FM4S 2024).
To accompany OpenPhenom-S/16, Recursion is releasing the RxRx3-core dataset, a challenge dataset in phenomics optimized for the research community. RxRx3-core includes labeled images of 735 genetic knockouts and 1,674 small-molecule perturbations drawn from the RxRx3 dataset, image embeddings computed with OpenPhenom-S/16, and associations between the included small molecules and genes. “Mapping the mechanisms by which drugs exert their actions is an important challenge in advancing the use of high-dimensional biological data like phenomics. We are excited to release the first dataset of this scale probing concentration-response along with a benchmark and model to enable the research community to rapidly advance this space,” commented Imran Haque, Ph.D., SVP of AI and Digital Sciences at Recursion.
To access OpenPhenom-S/16 on Google Cloud, through Hugging Face, please visit RxRx.ai/phenom. For more details on RxRx3-core, find the dataset at https://rxrx.ai/rxrx3-core. Please sign up at RxRx.ai to receive updates on all data and model releases from Recursion.
About Recursion
Recursion (NASDAQ: RXRX) is a clinical stage TechBio company leading the space by decoding biology to industrialize drug discovery. Enabling its mission is the Recursion OS, a platform built across diverse technologies that continuously expands one of the world’s largest proprietary biological and chemical datasets. Recursion leverages sophisticated machine-learning algorithms to distill from its dataset a collection of trillions of searchable relationships across biology and chemistry unconstrained by human bias. By commanding massive experimental scale — up to millions of wet lab experiments weekly — and massive computational scale — owning and operating one of the most powerful supercomputers in the world, Recursion is uniting technology, biology and chemistry to advance the future of medicine.
Recursion is headquartered in Salt Lake City, where it is a founding member of BioHive, the Utah life sciences industry collective. Recursion also has offices in Toronto, Montréal, London, and the San Francisco Bay Area. Learn more at www.Recursion.com, or connect on X (formerly Twitter) and LinkedIn.
Media Contact
Media@Recursion.com
Investor Contact
Investor@Recursion.comForward-Looking Statements
This document contains information that includes or is based upon "forward-looking statements" within the meaning of the Securities Litigation Reform Act of 1995, including, without limitation, those regarding the performance of the OpenPhenom model and its impact on the pharmaceutical industry, other life sciences companies, and drug discovery and other uses; and all other statements that are not historical facts. Forward-looking statements may or may not include identifying words such as “plan,” “will,” “expect,” “anticipate,” “intend,” “believe,” “potential,” “continue,” and similar terms. These statements are subject to known or unknown risks and uncertainties that could cause actual results to differ materially from those expressed or implied in such statements, including but not limited to: challenges inherent in pharmaceutical research and development, including the timing and results of preclinical and clinical programs, where the risk of failure is high and failure can occur at any stage prior to or after regulatory approval due to lack of sufficient efficacy, safety considerations, or other factors; our ability to leverage and enhance our drug discovery platform; our ability to obtain financing for development activities and other corporate purposes; the success of our collaboration activities; our ability to obtain regulatory approval of, and ultimately commercialize, drug candidates; our ability to obtain, maintain, and enforce intellectual property protections; cyberattacks or other disruptions to our technology systems; our ability to attract, motivate, and retain key employees and manage our growth; inflation and other macroeconomic issues; and other risks and uncertainties such as those described under the heading “Risk Factors” in our filings with the U.S. Securities and Exchange Commission, including our Annual Report on Form 10-K and Quarterly Report on Form 10-Q. All forward-looking statements are based on management’s current estimates, projections, and assumptions, and Recursion undertakes no obligation to correct or update any such statements, whether as a result of new information, future developments, or otherwise, except to the extent required by applicable law.
A photo accompanying this announcement is available at https://www.globenewswire.com/NewsRoom/AttachmentNg/4ab2ee8f-0ec0-4917-90f7-6aa9f09e1feb